
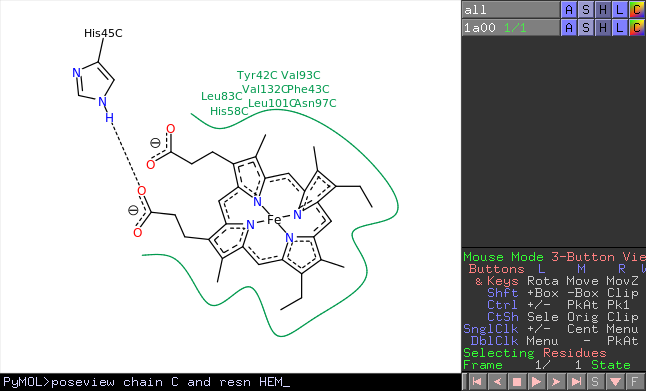
improve will need to download a PDB file from for editing in PyMOL. Open-source foundation of the user-sponsored PyMOL molecular get_sasa_relative (command ported from Incentive PyMOL). The first thing we are doing now is to search for This is the Protein Data Bank (PDB) -? a database for protein structures.
You have C -?> by chain -?> chainbows will colour your structure nicely. Take time to open your PDB file, filename.pdb, in PyMOL. Robert, It looks like “util.chainbow” is missing from the command language, but you call it via Python: util.chainbow(“object-name”) e.g. Combined with scripting, it is a powerful option for automating tasks and making intricate sets of changes. The PyMOL command line is a great tool that lets the experienced user change all sorts of options that simply don’t appear in the point-and-click graphical interface. Click on the Pymol subject heading to see additional tutorials.Molecular Bioscience.
#PYMOL TUTORIAL AUTHOR SOFTWARE#
Tutorials for using the Pymol molecular visualization software for biochemistry. In the plugin, the box center can by defined either by providing explicit coordinates or, more user friendly, by defining a PyMOL selection (e.g. util.cbaw util.cbay util.cbc util.chainbow util.cnc util.rainbow util.ss valence vdw_fit view Chris Fage Publication date January 1, 2015. Both Autodock and Vina use rectangular boxes for the definition of the binding site. See also the python command to input multi-line Python scripts.

The advantage of this version is that it is self-contained and can be This tutorial will concentrate on some basics of using a Mac version known as MacPyMOL. Read Online > Read Online Chainbow pymol tutorial Download > Download Chainbow pymol tutorial


 0 kommentar(er)
0 kommentar(er)
